
We can Blast these unmapped reads against phage databases and plasmid databases for species identification. When sequence alignment is finished, there may be a significant number of unmapped reads, which often represent inserted phage and/or plasmid sequences. These are commonly seen in bacterial sequences and in most cases represent bacteriophage, prophage, and plasmid insertions. This level of coverage often yields the uniform coverage seen in this example.Īlso, note the distinct gap around locus 4,100,000. Using tools like the Illumina Sequence Coverage Calculator, our own internal coverage calculators, and extensive experience with bacterial sequencing projects, we usually recommend a minimum of 100X coverage. Note the uniform coverage across the entire reference genome. = 22.6 reads and pairwise identity = 99.3%. coli sample was aligned to the reference strain with the Geneious Prime mapping algorithm. A total of 6,346,413 reads (DNA fragments) from the E. coli bacterial sample aligned to the reference genome E. We then perform sequence alignments of the FastQ files against the reference genomes. Our client researchers generally choose appropriate reference genomes for their projects, although we may assist them in identifying proper genomes. When sequencing runs are finished, our sequencers generate raw FastQ files, which typically contain millions of short DNA fragments (“reads”).
Geneious prime algorithm mac#
Last but not least, Geneious Prime is the perfect tool to have on your Mac if you need to easily insert DNA fragments into a vector with matching overhangs, to ligate multiple fragments with matching overhangs, to simulate restriction cloning, InFusion Cloning and Gibson Assembly, as well as conversional PCR and TOPO cloning.We often use Geneious for the alignment of sequenced samples to reference genomes. In addition, with the help of Geneious Prime you can highlight plasmids using terminators, common promoters, restriction sites, cloning sites, affinity tags, reporter genes, replication origins, open reading frames and selectable marker genes. Comprehensive biochemistry analysis tool with advanced molecular cloning Also, thanks to the PlasMapper you can automatically annotate plasmid maps and expression vectors. Moreover, Geneious Prime can help you design primers, simulate PCR and test cloning strategies using smart tools for Gibson and restriction cloning. You can even read arbitrarily large documents like NGS reads or contig assemblies. Hence, you can assemble multiple combinations of Sanger, 454, SOLiD and Illumina data at the same time.

Furthermore, Geneious Prime is capable to handle data from various types of sequencing machines with reads of different lengths, including paired-reads and mixed reads from different sequencing machines. The versatile Geneious Assemble is designed to manage read errors caused by incorrect bases or short indels. User-oriented graphical interface and trustworthy reference mapping

On top of that, you can enjoy the eye-catching genome browser, sequence assembly and reference mapping. Regardless of your data type, you can use Geneious Prime to quickly assemble it using fine-tuned and advanced algorithms.
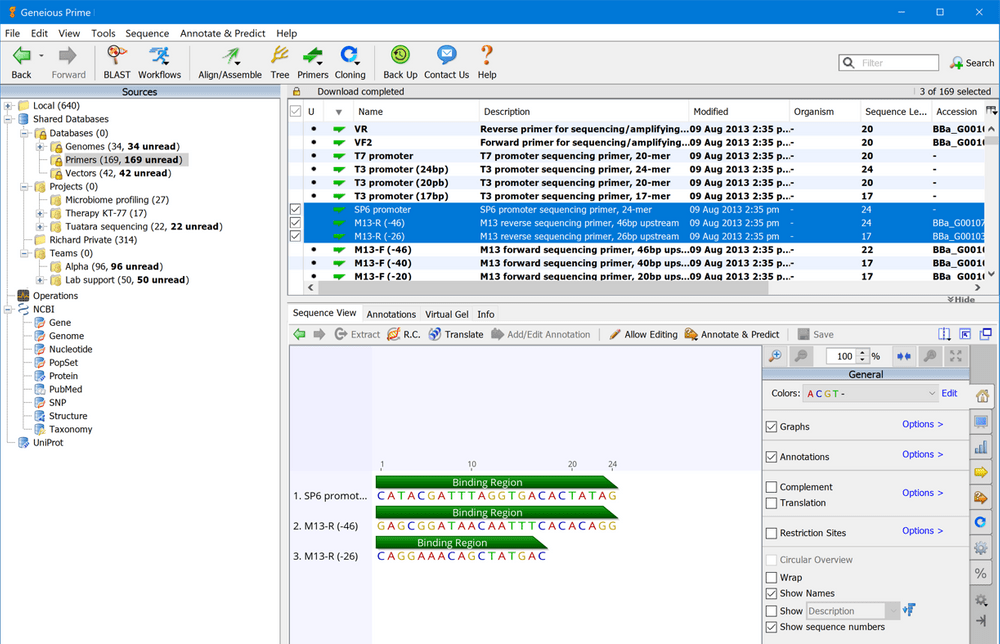
What is more, you can display phylogeny along with the alignment and link back to a tree, and use the highlighting feature to quickly locate disagreements.

You can also view annotations on the alignments, remove or insert gaps, strip columns, fix bad calls and join sequences. Thanks to the visual sequence alignment and editing you can effortlessly choose the desired algorithms to precisely align sequences without working with multiple files. Intuitive and easy-to-use sequence editing and analysis tools Geneious Prime is a well-designed and powerful sequence alignment, assembly and analysis application for genomics, microbiology, virology, infectious disease, synthetic biology, ecology, conservation genetics, population genetics, evolutionary biology, agriculture, as well as crop and plant science.


 0 kommentar(er)
0 kommentar(er)
